Trna Vs Mrna Vs Rrna
Eukaryotic Transcription
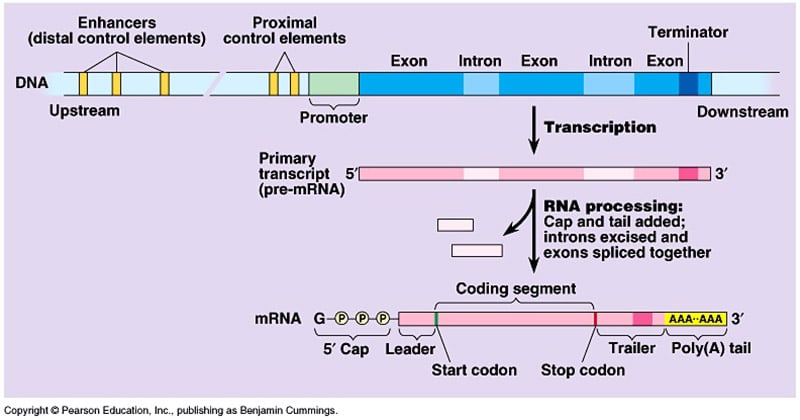
- Transcription is the process by which the information in a strand of Dna is copied into a new molecule of RNA.
- It is the first footstep of factor expression, in which a particular segment of Deoxyribonucleic acid is copied into RNA (especially mRNA) by the enzyme RNA polymerase.
- It results in a complementary, antiparallel RNA strand called a chief transcript.
Transcription in Eukaryotes
Transcription occurs in eukaryotes in a manner that is like to prokaryotes with reference to the bones steps involved. Yet, some major differences between them include:
- Initiation is more than complex.
- Termination does not involve stem-loop structures.
- Transcription is carried out by iii enzymes (RNA polymerases I, 2 and III).
- The regulation of transcription is more extensive than prokaryotes.
Enzyme(south) Involved in Eukaryotic Transcription
Different prokaryotes where all RNA is synthesized by a unmarried RNA polymerase, the nucleus of a eukaryotic cell has iii RNA polymerases responsible for transcribing different types of RNA.
- RNA polymerase I (RNA Pol I) is located in the nucleolus and transcribes the 28S, 18S, and 5.8S rRNA genes.
- RNA polymerase II (RNA Pol Ii) is located in the nucleoplasm and transcribes protein-coding genes, to yield pre-mRNA, and also the genes encoding small nucleolar RNAs (snoRNAs) involved in rRNA processing and small nuclear RNAs (snRNAs) involved in mRNA processing, except for U6 snRNA.
- RNA polymerase 3 (RNA Pol III) is too located in the nucleoplasm. It transcribes the genes for tRNA, 5S rRNA, U6 snRNA, and the 7S RNA associated with the indicate recognition particle (SRP) involved in the translocation of proteins across the endoplasmic reticulum membrane.
- Each of the 3 eukaryotic RNA polymerases contains 12 or more subunits so these are big complex enzymes.
- The genes encoding some of the subunits of each eukaryotic enzyme show DNA sequence similarities to genes encoding subunits of the cadre enzyme of E. coli RNA polymerase.
- However, four to seven other subunits of each eukaryotic RNA polymerase are unique in that they show no similarity either with bacterial RNA polymerase subunits or with the subunits of other eukaryotic RNA polymerases.
Features of Eukaryotic Transcription
- Transcription in eukaryotes occurs within the nucleus and mRNA moves out of the nucleus into the cytoplasm for translation.
- The initiation of RNA synthesis by RNA polymerase is directed past the presence of a promoter site on the 5' side of the transcriptional beginning site.
- The RNA polymerase transcribes 1 strand, the antisense (-) strand, of the Deoxyribonucleic acid template.
- RNA synthesis does not require a primer.
- RNA synthesis occurs in the 5' → three' direction with the RNA polymerase catalyzing a nucleophilic attack past the 3-OH of the growing RNA concatenation on the alpha-phosphorus atom on an incoming ribonucleoside five-triphosphate.
- mRNA in eukaryotes is processed from the chief RNA transcript, a procedure called maturation.
Process of Eukaryotic Transcription
The basic mechanism of RNA synthesis by these eukaryotic RNA polymerases can be divided into the following phases:
Initiation Phase
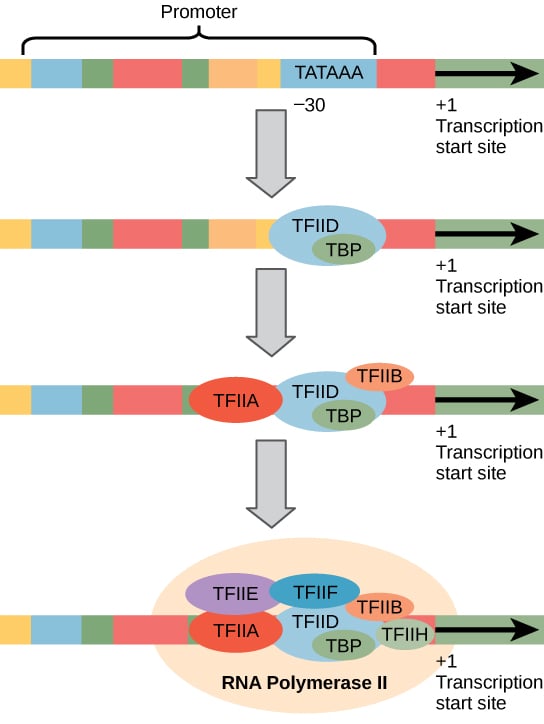
- During initiation, RNA polymerase recognizes a specific site on the Dna, upstream from the gene that will be transcribed, called a promoter site and so unwinds the DNA locally.
- Most promoter sites for RNA polymerase 2 include a highly conserved sequence located most 25–35 bp upstream (i.e. to the v side) of the start site which has the consensus TATA(A/T)A(A/T) and is called the TATA box.
- Since the start site is denoted as position +i, the TATA box position is said to be located at nearly position -25.
- The TATA box sequence resembles the -10 sequence in prokaryotes (TATAAT) except that it is located farther upstream.
- Both elements have essentially the same office, namely recognition by the RNA polymerase in order to position the enzyme at the correct location to initiate transcription.
- The sequence around the TATA box is too important in that it influences the efficiency of initiation. Transcription is also regulated by upstream command elements that prevarication 5′ to the TATA box.
- Some eukaryotic protein-coding genes lack a TATA box and accept an initiator element instead, centered around the transcriptional initiation site.
- In social club to initiate transcription, RNA polymerase II requires the assistance of several other proteins or protein complexes, called general (or basal) transcription factors, which must assemble into a circuitous on the promoter in order for RNA polymerase to bind and start transcription.
- These all have the generic name of TFII (for Transcription Cistron for RNA polymerase II).
- The first event in initiation is the binding of the transcription cistron IID (TFIID) protein complex to the TATA box via one its subunits chosen TBP (TATA box bounden poly peptide).
- Every bit presently equally the TFIID complex has jump, TFIIA binds and stabilizes the TFIID-TATA box interaction. Adjacent, TFIIB binds to TFIID.
- However, TFIIB tin also demark to RNA polymerase 2 and then acts every bit a bridging poly peptide. Thus,
- RNA polymerase Ii, which has already complexed with TFIIF, now binds.
- This is followed by the binding of TFIIE and H. This final poly peptide complex contains at least 40 polypeptides and is called the transcription initiation circuitous.
- Those poly peptide-coding genes that have an initiator element instead of a TATA box appear to need another protein(due south) that binds to the initiator element.
- The other transcription factors then bind to form the transcription initiation complex in a like manner to that described to a higher place for genes possessing a TATA box promoter.
Elongation Phase
TFIIH has two functions:
- It is a helicase, which means that information technology tin use ATP to unwind the Deoxyribonucleic acid helix, allowing transcription to begin.
- In add-on, it phosphorylates RNA polymerase 2 which causes this enzyme to change its conformation and dissociate from other proteins in the initiation complex.
- The key phosphorylation occurs on a long C-last tail chosen the C-concluding domain (CTD) of the RNA polymerase Two molecule.
- Interestingly, simply RNA polymerase Two that has a non-phosphorylated CTD can initiate transcription but only an RNA polymerase II with a phosphorylated CTD can elongate RNA.
- RNA polymerase II now starts moving forth the Deoxyribonucleic acid template, synthesizing RNA, that is, the process enters the elongation phase.
- RNA synthesis occurs in the 5' → three' direction with the RNA polymerase catalyzing a nucleophilic set on past the 3-OH of the growing RNA chain on the alpha-phosphorus atom on an incoming ribonucleoside five-triphosphate.
- The RNA molecule made from a protein-coding gene past RNA polymerase II is chosen a primary transcript.
Termination Phase
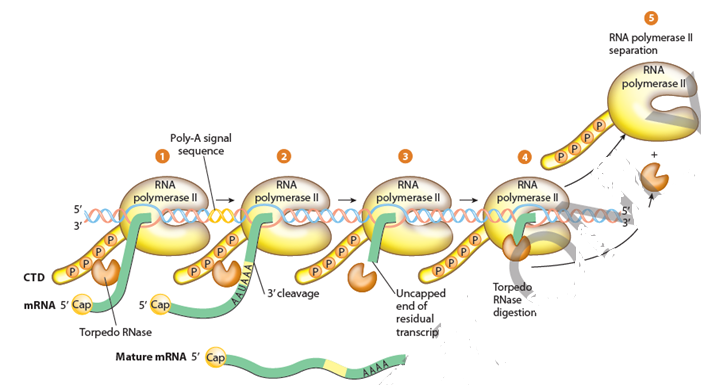
- Elongation of the RNA chain continues until termination occurs.
- Unlike RNA polymerase in prokaryotes, RNA polymerase Two does non terminate transcription at a specific site only rather transcription tin can stop at varying distances downstream of the gene.
- RNA genes transcribed by RNA Polymerse Ii lack any specific signals or sequences that direct RNA Polymerase Ii to stop at specific locations.
- RNA Polymerase II can continue to transcribe RNA anywhere from a few bp to thousands of bp past the actual finish of the factor.
- The transcript is cleaved at an internal site before RNA Polymerase II finishes transcribing. This releases the upstream portion of the transcript, which will serve as the initial RNA prior to further processing (the pre-mRNA in the case of protein-encoding genes.)
- This cleavage site is considered the "end" of the gene. The remainder of the transcript is digested past a 5′-exonuclease (called Xrn2 in humans) while it is still beingness transcribed by the RNA Polymerase Two.
- When the 5′-exonulease "catches up" to RNA Polymerase II by digesting abroad all the overhanging RNA, it helps undo the polymerase from its DNA template strand, finally terminating that circular of transcription.
RNA processing
The primary eukaryotic mRNA transcript is much longer and localised into the nucleus, when it is also called heterogenous nuclear RNA (hnRNA) or pre- mRNA.
It undergoes various processing steps to alter into a mature RNA:
Cleavage
- Larger RNA precursors are cleaved to form smaller RNAs.
- Master transcript is cleaved past ribonuclease-P (an RNA enzyme) to form 5-vii tRNA precursors.
Capping and Tailing
- Initially at the 5′ cease a cap (consisting of 7-methyl guanosine or 7 mG) and a tail of poly A at the three′ cease are added.
- The cap is a chemically modified molecule of guanosine triphosphate (GTP).
Splicing
- The eukaryotic primary mRNAs are fabricated up of two types of segments; non-coding introns and the coding exons.
- The introns are removed by a process called RNA splicing where ATP is used to cut the RNA, releasing the introns and joining two side by side exons to produce mature mRNA.
Nucleotide Modifications
- They are most common in tRNA-methylation (e.g., methyl cytosine, methyl guanosine), deamination (e.chiliad., inosine from adenine), dihydrouracil, pseudouracil, etc.
Mail service-transcription processing is required to convert chief transcript into functional RNAs.
Significance
- Transcription of Dna is the method for regulating gene expression.
- It occurs in preparation for and is necessary for protein translation.
References
- David Hames and Nigel Hooper (2005). Biochemistry. 3rd ed. Taylor & Francis Group: New York.
- Bailey, W. R., Scott, Due east. K., Finegold, S. M., & Baron, E. J. (1986). Bailey and Scott'southward Diagnostic microbiology. St. Louis: Mosby.
- Madigan, M. T., Martinko, J. M., Bender, Thousand. S., Buckley, D. H., & Stahl, D. A. (2015). Brock biological science of microorganisms (Fourteenth edition.). Boston: Pearson.
- http://world wide web.biologydiscussion.com/rna/transcription/transcription-in-prokaryotes-and-eukaryotes-with-diagram/15546
- https://courses.lumenlearning.com/boundless-biology/chapter/eukaryotic-transcription/
Eukaryotic Transcription
Trna Vs Mrna Vs Rrna,
Source: https://microbenotes.com/eukaryotic-transcription/
Posted by: mirelesbobst1939.blogspot.com


0 Response to "Trna Vs Mrna Vs Rrna"
Post a Comment